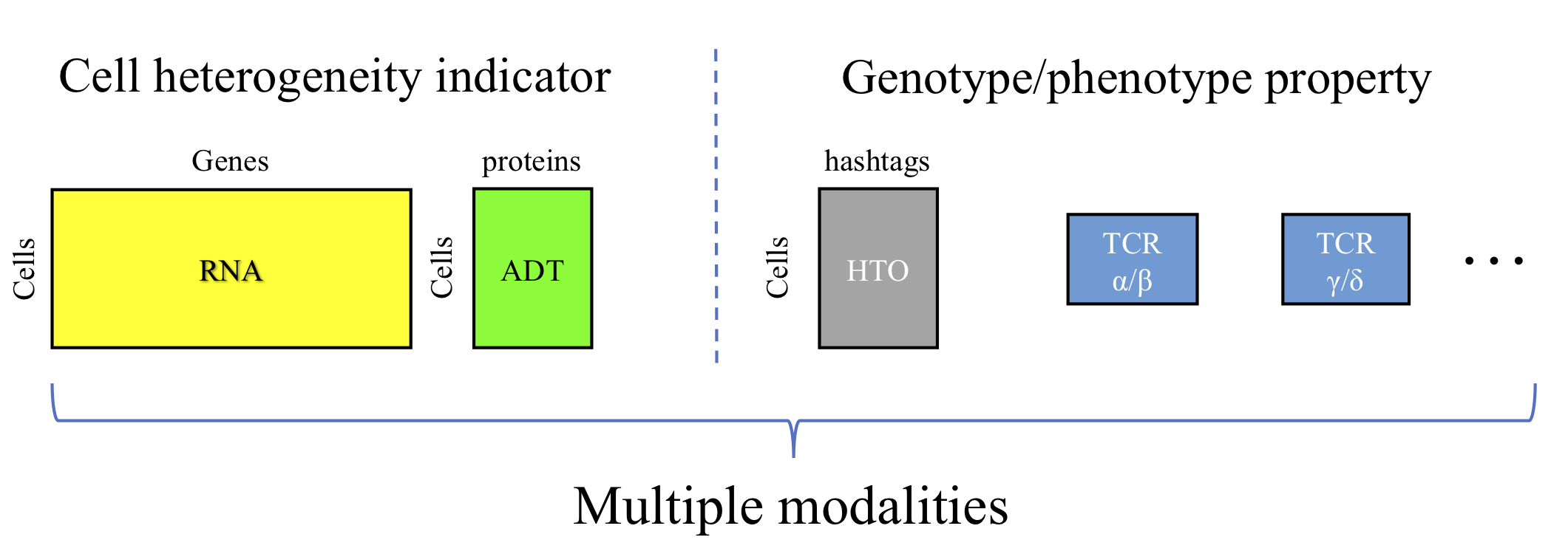
Example datasets and Rmd scripts
| Dataset (Click to download) | Rmd file (Click to download) |
|---|---|
10X 1K  |
10X1K.Rmd  |
10X 5K  |
10X5K.Rmd  |
10X 10K  |
10X10K.Rmd  |
10X 10K MALT  |
10X10Kmalt.Rmd  |
RNA  ADT
ADT 
|
CiteSeq.Rmd  |
Load data from multiple resources (Click for details)
Users are allowed to load data from multiple resources. We adopted data object strcuture desgin and pre-processing steps from Seurat V3. Users famliar with Seurat can use Seurat functions to load data and create objects. We also provided some functions for data loading.
Joint analysis achieves precise cell clustering by integrating both transcriptome and surface protein signals (Click for details)
Using “LinQ-View”, we are able to perform joint analysis for multi-modal datasets and compare the results with transcriptome or surface protein alone. We performed joint analysis on several public multi-modal datasets (see method section for details). Results showed joint analysis achieves better clustering quality and deeper resolutions.
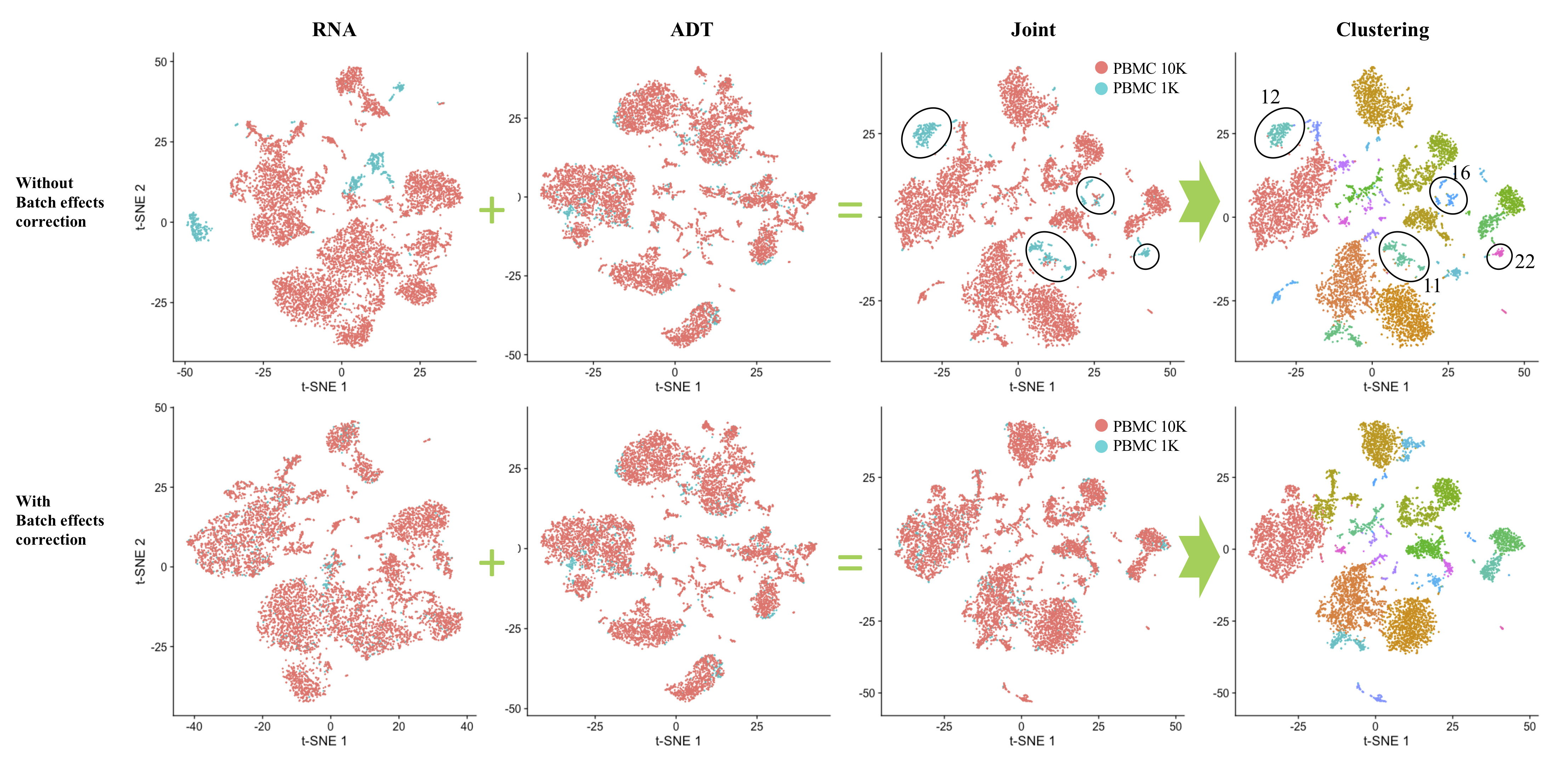
Batch effects correction for multi-modal data with joint analysis (Click for details)
To evaluate the compatibility of “LinQ-View” with conventional computational batch effects correction methods, we performed joint analysis using “LinQ-View” on two datasets of PBMCs from healthy donors (called PBMC 1K and PBMC 10K). We performed joint analysis on two independent runs: one with batch effects correction and one without as control. Here, we use Seurat V3 to remove the batch effects.